Host-Pathogen interactions
Our Focus
01
Development of CRISPR
screening technologies
02
Bacterial toxins and
other virulence factors
03
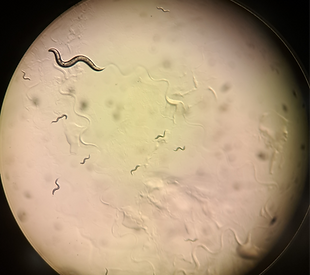
Bacteria-nematode
interactions


Using existing and developing novel CRISPR-based screening approaches to understand how bacteria invade hosts and cause disease
Elucidating mechanisms of bacterial toxin and virulence factor interactions with and within host cells
Understanding how certain bacteria colonize nematode guts and, in some cases, kill them
We let our curiosity guide us to better understand nature, while striving to make a positive impact on society
Our Team
Our Papers
-
Hart, S. K., Müller, S., Wessels, H. H., Méndez-Mancilla, A., Drabavicius, G., Choi, O., & Sanjana, N. E. (2025). Precise RNA targeting with CRISPR–Cas13d. Nature Biotechnology, 1-6.
-
Drabavicius, G. & D. Daelemans (2021). Intermedilysin cytolytic activity depends on heparan sulfates and membrane composition PLoS genetics, 17(2), e1009387.
-
Wilkinson, M., Drabavicius, G., Silanskas, A., Gasiunas, G., Siksnys, V., & Wigley, D. B. (2019). Structure of the DNA-bound spacer capture complex of a type II CRISPR-Cas system. Molecular cell, 75(1), 90-101.
-
Drabavicius, G., Sinkunas, T., Silanskas, A., Gasiunas, G., Venclovas, Č., & Siksnys, V. (2018). DnaQ exonuclease‐like domain of Cas2 promotes spacer integration in a type I‐E CRISPR‐Cas system. EMBO reports 19(7), e45543
The Drab Lab papers
Working on it
Pre-Drab Lab papers
Contact Us
Address
Medical Sciences Center
Žaliųjų ežerų g. 2
LT - 08422, Vilnius, Lithuania
Contact
+370-617-15238





